- Joined
- Jun 20, 2016
- Messages
- 264
- Reaction score
- 48
I understand the question in general but I was wondering if someone could explain what technique D is.
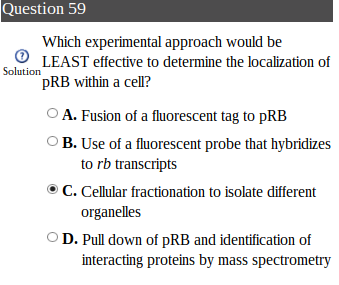
What does it mean by pull down?
I am assuming this is effective because certain proteins (identified by mass spectrometry) is only localized at certain places in the cell?
What does it mean by pull down?
I am assuming this is effective because certain proteins (identified by mass spectrometry) is only localized at certain places in the cell?
