Hey guys!
I'm currently stuck on this question that I posted below.
How can you analyze the "rate" of histone acetylation by western blot?
From my knowledge, I thought Western blot just shows you whether the protein is there or not and the relative amounts (compared to control).
Why couldn't RT-PCR detect levels of expression? The increase in HA means an increase in gene expression, which RT-PCR analyzes as well.
Thank you!
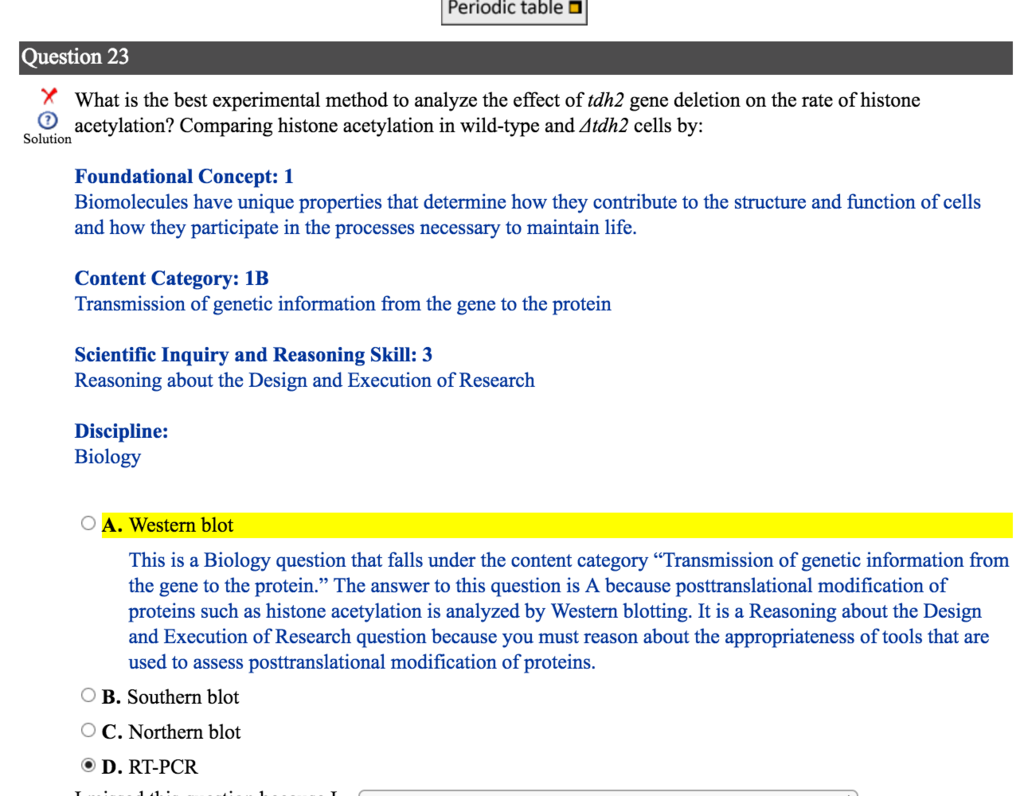
I'm currently stuck on this question that I posted below.
How can you analyze the "rate" of histone acetylation by western blot?
From my knowledge, I thought Western blot just shows you whether the protein is there or not and the relative amounts (compared to control).
Why couldn't RT-PCR detect levels of expression? The increase in HA means an increase in gene expression, which RT-PCR analyzes as well.
Thank you!
